Data inside DNA, or the world inside a shoe box
Back in 2016, a post signed by Thomas Barnet Jr. titled “The Zettabyte Era Officially Begins” appeared on Cisco Blog. What’s all about?
The post was referring to the global Internet traffic as measured by Cisco, which in 2016 had just exceeded the ZB1, and expected to exceed the 3 ZB by 2021. But the traffic is still nothing compared to the generated data (which exceeded the ZB already in 2012), whereas IDC, in its report Data Age 2025 showed that the threshold of 20 ZB was already exceeded this year and that this exponential growth would lead to breaking through the 160 ZB by 2025!

A deluge of data
We are generating an immense amount of data, and we are rapidly reaching the capacity limit of the current technology to handle it. Some might argue that a large part of the data generated is garbage that could easily be deleted without any problem, but it is difficult to understand today what might become relevant in the future, so this can certainly not be considered a solution.
Big Data is already a challenge in terms of computing capacity today, but it will soon become a challenge in terms of space with today’s technologies: SSD media have brought some performance improvement over magnetic hard disks, but for what concerns long-term storage we are still stuck with magnetic tapes.
Genetics to the rescue?
In 2007, GM Skinner, K. Visscher, and M. Mansuripur published a fairly revolutionary paper in the Journal of Bionanoscience, titled Biocompatible Writing of Data into DNA, where they used a simple DNA-based storage scheme. In this work, the group demonstrated the possibility to “write” information in DNA strands and to read it using a specific gel. The method was still rudimentary but the way was paved.

Sequencing and Synthesis
The process of reading DNA, better known as “sequencing”, received a major boost from the work of the NHGRI within the scope of the Human Genome Project, which was completed in 2003.

The DNA is made of 4 bases: Adenine, Guanine, Thymine, and Cytosine. The “trick” is that the only combinations allowed are between Adenine and Thymine, and between Cytosine and
Why DNA?
There are many advantages:
- Density: DNA is above all incredibly dense. Already last year the threshold of 200 PetaBytes (1000 TB) per gram was exceeded. It is estimated that all data on the Internet today could be easily contained on DNA in the space of a shoe box (!).
- Loyalty: data recovery can be virtually error-free due to the accuracy of DNA replication methods.
- Sustainability: the energy required to maintain DNA-encoded information is a small fraction of that required by modern data centers.
- Longevity: DNA is a stable molecule that can last for thousands of years without degrading.
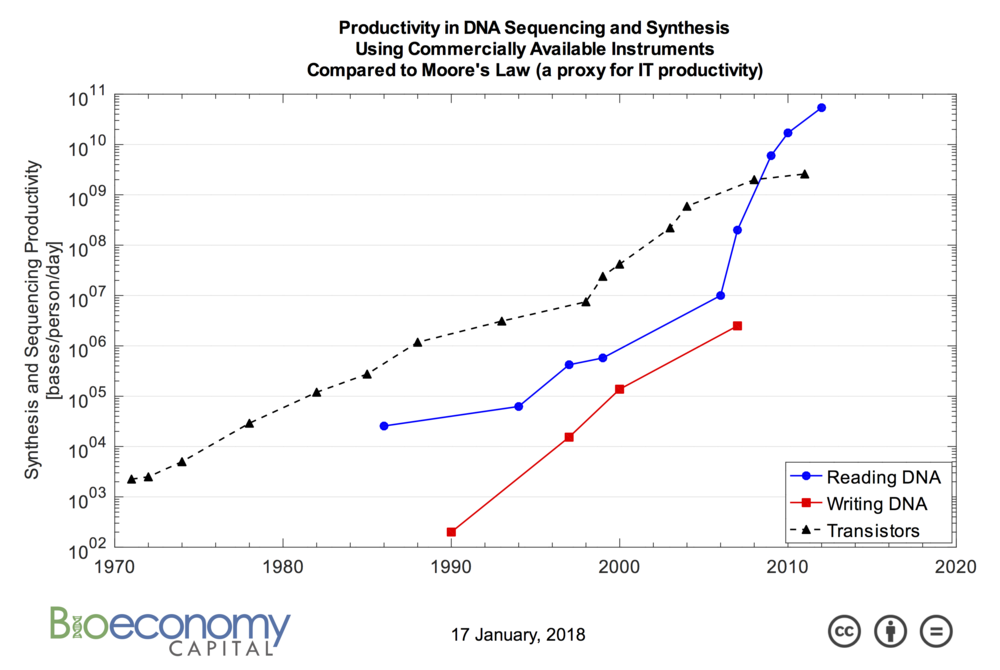
The sequencing technologies are now very advanced, and today there are even USB pocket sequencers (see below), and the most advanced devices allow the execution of many runs in parallel.

The writing (or synthesis) of DNA instead requires to “attach” together one base after another in a controlled environment, a very slow chemical process that dates back to 1981. However, given the huge market demand, there are companies like Twist Bioscience and DNA Script that have developed innovative synthesis technologies, based respectively on silicon and enzymatic synthesis, which promise volumes orders of magnitude higher than traditional ones. Moreover, just recently, two researchers at the Synthetic Biology Informatics department of JBEI presented a new synthesis methodology that could lead to the creation of 3D printers of DNA.
Since the work of Skinner & coll. the research has made huge progress: in 2015, Microsoft and MISL of the University of Washington created the DNA Storage project, establishing a record in 2016 by storing and successfully recovering 200 MB in strands of DNA. In 2017, in another important work, Y. Erlich and D. Zielinski, stored and recovered 2 MB of material with a density of over 200 PetaByte per gram, touching the theoretical limit postulated by Shannon, through the use of “fountain codes”.

To this day, the DNA synthesis/sequencing process is still expensive (we are talking about a few thousand dollars per MB in writing and 200 for reading) but this is bound to fall, both in view of the rapid evolution of the sector, due to the explosive request of engineered DNA, both because for the storage of the data it is possible to use ad-hoc synthesized DNA instead of the biological one. In this regard, it is expected that the extensive use of editing technologies such as CRISPR/Cas9, TALEN
Applications
The use of DNA for digitization is therefore not something that belongs to science fiction, but we are already starting to see the first prototypes of applications.
- Encryption:
Carverr , an American startup has developed a method to encrypt data into DNA molecules and offers a DNA-based password encryption service for $1,000. - Cloud: just last March Microsoft published a paper on Nature where it demonstrated the ability to perform DNA readings through
random access, dramatically increasing the efficiency of the sequencing process. Thanks to progress like this and those mentioned above, Microsoft seems to be starting to consider DNA for cloud backup for the future and is actively collaborating with Twist Biosciences. The costs are still very high but people at Redmond are convinced that this obstacle will easily be overcome if there is sufficient demand from the computer industry.
Note
One zettabyte is equivalent to about one billion terabytes (TB). If we consider that 1 TB is more or less the size of an average hard disk today, it is easy to realize the size of this traffic.
A fountain code is a way of taking data (eg a file) and transforming it into an actually unlimited number of coded chunks, so that the original file can be reassembled by any set of those pieces, as long as the total is slightly higher than the original size. What makes this type of algorithm remarkable is that it allows you to send information through “noisy” channels without requiring the receiver to send feedback on missing packets. In other words, having a file of 10 MB, for the recipient will suffice to
With Random Access in IT we mean the ability to access any location of the media without having to pass thru the previous locations (serial access).
Links
An Interactive Timeline of the Human Genome
Wikipedia: DNA Digital Storage
Storage
THE RISE OF DNA DATA STORAGE
Random access in large-scale DNA data storage
DNA data storage closer to becoming reality
Microsoft and University of Washington researchers set record for DNA storage
How DNA could store all the world’s data
Storing data in DNA brings nature into the digital universe
Toward practical high-capacity low-maintenance storage of digital information in synthesised DNA (pdf)
DNA storage: a new method for storing digital information
Will synthetic DNA push out Ledger and Trezor from the market?
Synthesis and Sequencing
DNA EXTRACTION WITH A 3D-PRINTED CENTRIFUGE
REVERSE ENGINEERING A DNA SEQUENCER
New Research Could Lead to DNA 3D Printer
DNA Fountain enables a robust and efficient storage architecture (pdf)
MinION: A complete DNA sequencer on a USB stick
DNA Sequencer Market: Emerging Industries, Potential Revenue, Cost Structure Analysis and Top Key Players
Applications
Bitcoin fanatics are storing their cryptocurrency passwords in DNA
3D Printing May Be the Key to Affordable Data Storage Using DNA
Damn Cool Algorithms: Fountain Codes
Andrea has been working in IT for almost 20 years, covering about everything, from development to business analysis, to project management.
Today we can tell he is a carefree gnome, passionate with Neurosciences, Artificial Intelligence, and photography







Leave a Comment